Nuclei Segmentation by StarDist¶
VisiumHD represents a revolutionary technology, potentially as impactful as the introduction of droplet-based single-cell methods. By eliminating gaps between spots, VisiumHD enables continuous and detailed tissue mapping, offering a subcellular resolution of 2µm. This resolution greatly improving the precision and accuracy of cellular reconstructions. As the technology is still in its early development stages, it is crucial to develop strategies that fully exploit the data it provides.
We introduce SMURF (Segmentation and Manifold UnRolling Framework) to leverage soft segmentation with VisiumHD data, facilitating the creation of a cells*genes anndata object. SMURF uses high-resolution images from VisiumHD for nuclei segmentation.
In our demonstration, we use StarDist for segmentation. Please install StarDist beforehand. Most segmentation tools rely on deep learning packages and require specific configurations. However, for this tutorial, there is no need to install any SMURF tools. We recommend users create a new environment specifically for segmentation. Researchers are encouraged to use their preferred segmentation methods. The segmentation results should be saved as a 2D NumPy array with the same dimensions as the input image. In this array, regions without nuclei should be labeled as ``0``, while nucleated regions should be assigned ``unique cell IDs``.
[1]:
import numpy as np
import matplotlib.pyplot as plt
import gzip
[2]:
data_path = 'smurf_example_data/'
save_path = 'results_example_mousebrain/'
Dataset Information¶
Access the dataset here: Visium HD CytAssist Gene Expression Libraries of Mouse Brain
Please input image_to_segmentation.npy.gz from the tutorial output.
[3]:
with gzip.GzipFile(data_path + 'image_to_segmentation.npy.gz', 'r') as f:
image = np.load(f)
Due to the large size of the image, we divide it into multiple parts. Here, the loop indicates the size of each segment. In other words, we will process the segmentation loop by loop, one segment at a time, with each neighboring segment overlapping by a gap size to facilitate correction.
[4]:
i_max = image.shape[0]
j_max = image.shape[1]
num = 0
loop = 4700
segmentation_results = {}
gap = 80
[5]:
# Import the StarDist 2D segmentation models.
# Import the recommended normalization technique for stardist.
from csbdeep.utils import normalize
from stardist.models import StarDist2D
# Import squidpy and additional packages needed for this tutorial.
import squidpy as sq
2024-11-30 09:34:34.115885: I tensorflow/core/util/port.cc:113] oneDNN custom operations are on. You may see slightly different numerical results due to floating-point round-off errors from different computation orders. To turn them off, set the environment variable `TF_ENABLE_ONEDNN_OPTS=0`.
2024-11-30 09:34:34.146737: E external/local_xla/xla/stream_executor/cuda/cuda_dnn.cc:9261] Unable to register cuDNN factory: Attempting to register factory for plugin cuDNN when one has already been registered
2024-11-30 09:34:34.146762: E external/local_xla/xla/stream_executor/cuda/cuda_fft.cc:607] Unable to register cuFFT factory: Attempting to register factory for plugin cuFFT when one has already been registered
2024-11-30 09:34:34.147672: E external/local_xla/xla/stream_executor/cuda/cuda_blas.cc:1515] Unable to register cuBLAS factory: Attempting to register factory for plugin cuBLAS when one has already been registered
2024-11-30 09:34:34.152938: I tensorflow/core/platform/cpu_feature_guard.cc:182] This TensorFlow binary is optimized to use available CPU instructions in performance-critical operations.
To enable the following instructions: AVX2 AVX512F AVX512_VNNI FMA, in other operations, rebuild TensorFlow with the appropriate compiler flags.
2024-11-30 09:34:34.807242: W tensorflow/compiler/tf2tensorrt/utils/py_utils.cc:38] TF-TRT Warning: Could not find TensorRT
_compat.py (124): The Shapely GEOS version (3.11.3-CAPI-1.17.3) is incompatible with the GEOS version PyGEOS was compiled with (3.10.4-CAPI-1.16.2). Conversions between both will be slow.
__init__.py (11): Geopandas was set to use PyGEOS, changing to shapely 2.0 with:
geopandas.options.use_pygeos = True
If you intended to use PyGEOS, set the option to False.
utils.py (369): pandas.Int64Index is deprecated and will be removed from pandas in a future version. Use pandas.Index with the appropriate dtype instead.
utils.py (369): pandas.Float64Index is deprecated and will be removed from pandas in a future version. Use pandas.Index with the appropriate dtype instead.
utils.py (369): pandas.UInt64Index is deprecated and will be removed from pandas in a future version. Use pandas.Index with the appropriate dtype instead.
[6]:
StarDist2D.from_pretrained("2D_versatile_he")
def stardist_2D_versatile_he(img, nms_thresh=None, prob_thresh=None):
# axis_norm = (0,1) # normalize channels independently
axis_norm = (0, 1, 2) # normalize channels jointly
# Make sure to normalize the input image beforehand or supply a normalizer to the prediction function.
# this is the default normalizer noted in StarDist examples.
img = normalize(img, 1, 99.8, axis=axis_norm)
model = StarDist2D.from_pretrained("2D_versatile_he")
labels, _ = model.predict_instances(
img, nms_thresh=nms_thresh, prob_thresh=prob_thresh
)
return labels
Found model '2D_versatile_he' for 'StarDist2D'.
2024-11-30 09:34:39.378100: I tensorflow/core/common_runtime/gpu/gpu_device.cc:1929] Created device /job:localhost/replica:0/task:0/device:GPU:0 with 22272 MB memory: -> device: 0, name: NVIDIA GeForce RTX 3090, pci bus id: 0000:65:00.0, compute capability: 8.6
Loading network weights from 'weights_best.h5'.
Loading thresholds from 'thresholds.json'.
Using default values: prob_thresh=0.692478, nms_thresh=0.3.
Since this HE image is contaminated, we have defined a function to clean the affected areas. There is no need to apply this function to your own image.
[7]:
import copy
def clean(array_im):
target_colors = np.array([[155, 60, 126],
[128,24,101],
[190,118,172],
[220,95,176],
[139,42,110],
[132,39,102],
[136,44,108],
[125,53,110],
[109,39,91],
[178,90,144],
[149,67,122],
[191,70,159],
[186,69,150 ],
[176,40,132 ]]
)
new_color = [250,139,209]
threshold = 20
for target_color in target_colors:
distances = np.sqrt(((array_im - target_color)**2).sum(axis=-1))
array_im[distances < threshold] = new_color
return array_im
[8]:
i= loop*0
j = loop*0
a_t = image[max(0,i-gap):min(i+loop,i_max),max(0,j-gap):min(j+loop,j_max)]
a = sq.im.ImageContainer(img=a_t, layer='image', lazy=True, scale=1.0, )
sq.im.segment(
img=a,
layer="image",
channel=None,
method=stardist_2D_versatile_he,
layer_added="segmented_stardist",
prob_thresh=0.02,
nms_thresh=None,
)
fig, axes = plt.subplots(1, 2, figsize=(20, 10))
a.show("image", ax=axes[0])
_ = axes[0].set_title("H&H")
a.show("segmented_stardist", cmap="jet", interpolation="none", ax=axes[1])
_ = axes[1].set_title("segmentation")
segmentation_results[(i,j)] = np.array(a['segmented_stardist'][:,:,0,0])
Found model '2D_versatile_he' for 'StarDist2D'.
Loading network weights from 'weights_best.h5'.
Loading thresholds from 'thresholds.json'.
Using default values: prob_thresh=0.692478, nms_thresh=0.3.
2024-11-30 09:34:41.630608: I external/local_xla/xla/stream_executor/cuda/cuda_dnn.cc:454] Loaded cuDNN version 8904
2024-11-30 09:34:41.733600: I external/local_tsl/tsl/platform/default/subprocess.cc:304] Start cannot spawn child process: No such file or directory
2024-11-30 09:34:44.910268: I external/local_tsl/tsl/platform/default/subprocess.cc:304] Start cannot spawn child process: No such file or directory

[9]:
i= loop*0
j = loop*1
a_t = image[max(0,i-gap):min(i+loop,i_max),max(0,j-gap):min(j+loop,j_max)]
a_t = clean(a_t) # No need to clean for your own data
a = sq.im.ImageContainer(img=a_t, layer='image', lazy=True, scale=1.0, )
sq.im.segment(
img=a,
layer="image",
channel=None,
method=stardist_2D_versatile_he,
layer_added="segmented_stardist",
prob_thresh=0.01,
nms_thresh=None,
)
fig, axes = plt.subplots(1, 2, figsize=(20, 10))
a.show("image", ax=axes[0])
_ = axes[0].set_title("H&H")
a.show("segmented_stardist", cmap="jet", interpolation="none", ax=axes[1])
_ = axes[1].set_title("segmentation")
segmentation_results[(i,j)] = np.array(a['segmented_stardist'][:,:,0,0])
Found model '2D_versatile_he' for 'StarDist2D'.
Loading network weights from 'weights_best.h5'.
Loading thresholds from 'thresholds.json'.
Using default values: prob_thresh=0.692478, nms_thresh=0.3.
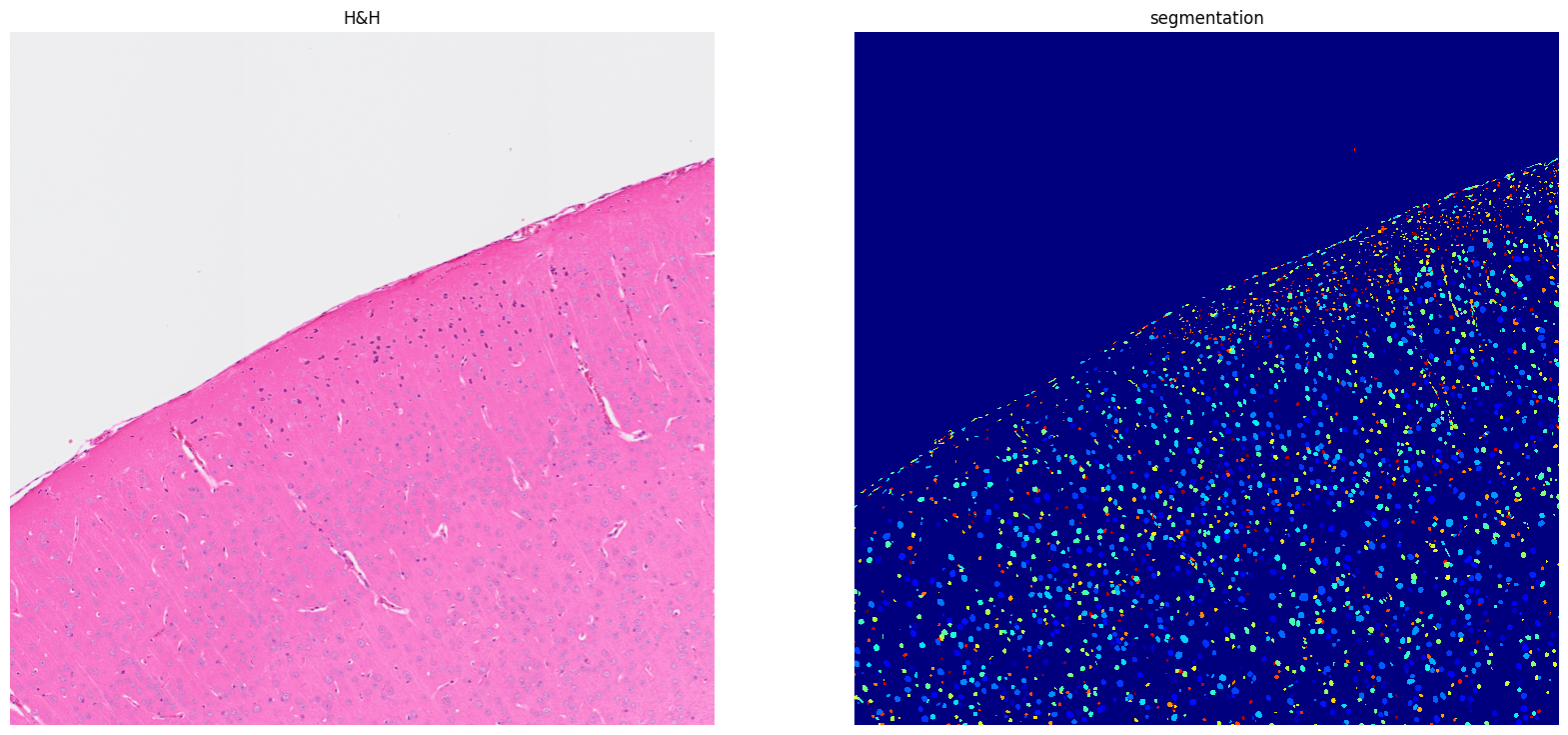
[10]:
i= loop*0
j = loop*2
a_t = image[max(0,i-gap):min(i+loop,i_max),max(0,j-gap):min(j+loop,j_max)]
a_t = clean(a_t) # No need to clean for your own data
a = sq.im.ImageContainer(img=a_t, layer='image', lazy=True, scale=1.0, )
sq.im.segment(
img=a,
layer="image",
channel=None,
method=stardist_2D_versatile_he,
layer_added="segmented_stardist",
prob_thresh=0.01,
nms_thresh=None,
)
fig, axes = plt.subplots(1, 2, figsize=(20, 10))
a.show("image", ax=axes[0])
_ = axes[0].set_title("H&H")
a.show("segmented_stardist", cmap="jet", interpolation="none", ax=axes[1])
_ = axes[1].set_title("segmentation")
segmentation_results[(i,j)] = np.array(a['segmented_stardist'][:,:,0,0])
Found model '2D_versatile_he' for 'StarDist2D'.
Loading network weights from 'weights_best.h5'.
Loading thresholds from 'thresholds.json'.
Using default values: prob_thresh=0.692478, nms_thresh=0.3.
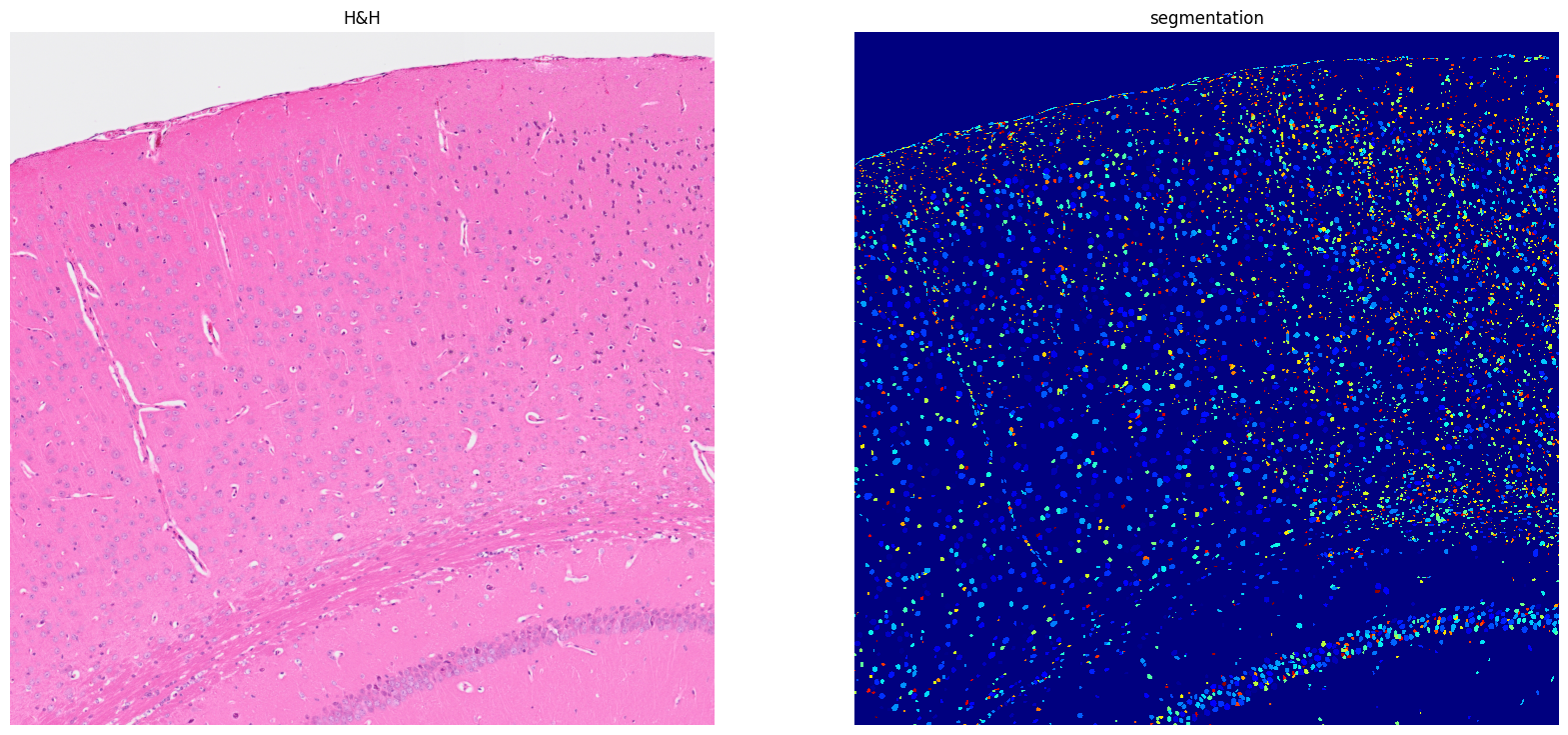
[11]:
i= loop*0
j = loop*3
a_t = image[max(0,i-gap):min(i+loop,i_max),max(0,j-gap):min(j+loop,j_max)]
a_t = clean(a_t) # No need to clean for your own data
a = sq.im.ImageContainer(img=a_t, layer='image', lazy=True, scale=1.0, )
sq.im.segment(
img=a,
layer="image",
channel=None,
method=stardist_2D_versatile_he,
layer_added="segmented_stardist",
prob_thresh=0.03,
nms_thresh=None,
)
fig, axes = plt.subplots(1, 2, figsize=(20, 10))
a.show("image", ax=axes[0])
_ = axes[0].set_title("H&H")
a.show("segmented_stardist", cmap="jet", interpolation="none", ax=axes[1])
_ = axes[1].set_title("segmentation")
segmentation_results[(i,j)] = np.array(a['segmented_stardist'][:,:,0,0])
Found model '2D_versatile_he' for 'StarDist2D'.
Loading network weights from 'weights_best.h5'.
Loading thresholds from 'thresholds.json'.
Using default values: prob_thresh=0.692478, nms_thresh=0.3.
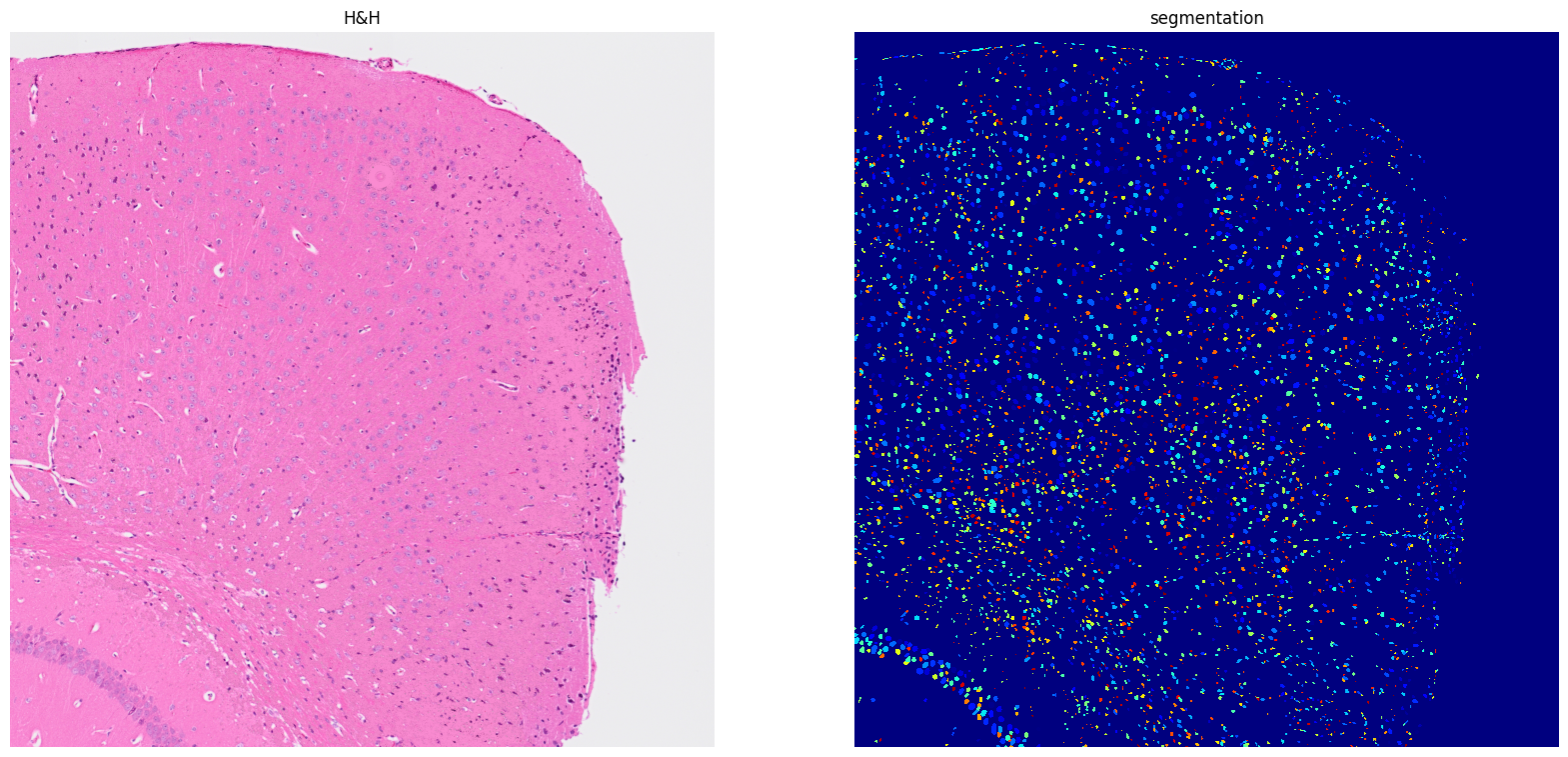
[12]:
i= loop*1
j = loop*0
a_t = image[max(0,i-gap):min(i+loop,i_max),max(0,j-gap):min(j+loop,j_max)]
a = sq.im.ImageContainer(img=a_t, layer='image', lazy=True, scale=1.0, )
sq.im.segment(
img=a,
layer="image",
channel=None,
method=stardist_2D_versatile_he,
layer_added="segmented_stardist",
prob_thresh=0.01,
nms_thresh=None,
)
fig, axes = plt.subplots(1, 2, figsize=(20, 10))
a.show("image", ax=axes[0])
_ = axes[0].set_title("H&H")
a.show("segmented_stardist", cmap="jet", interpolation="none", ax=axes[1])
_ = axes[1].set_title("segmentation")
segmentation_results[(i,j)] = np.array(a['segmented_stardist'][:,:,0,0])
Found model '2D_versatile_he' for 'StarDist2D'.
Loading network weights from 'weights_best.h5'.
Loading thresholds from 'thresholds.json'.
Using default values: prob_thresh=0.692478, nms_thresh=0.3.
WARNING:tensorflow:5 out of the last 5 calls to <function Model.make_predict_function.<locals>.predict_function at 0x7fb049e43550> triggered tf.function retracing. Tracing is expensive and the excessive number of tracings could be due to (1) creating @tf.function repeatedly in a loop, (2) passing tensors with different shapes, (3) passing Python objects instead of tensors. For (1), please define your @tf.function outside of the loop. For (2), @tf.function has reduce_retracing=True option that can avoid unnecessary retracing. For (3), please refer to https://www.tensorflow.org/guide/function#controlling_retracing and https://www.tensorflow.org/api_docs/python/tf/function for more details.
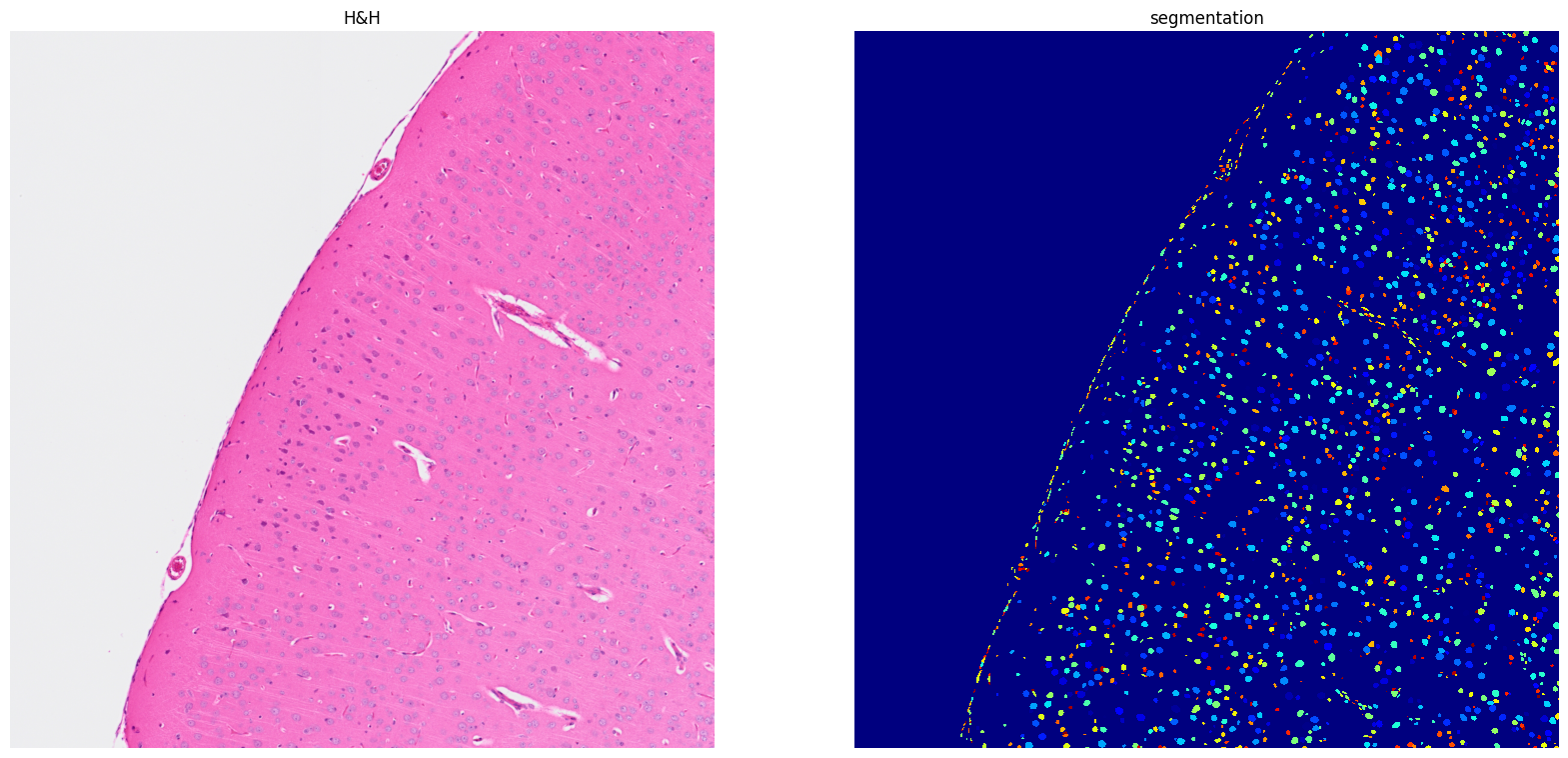
[13]:
i= loop*1
j = loop*1
a_t = image[max(0,i-gap):min(i+loop,i_max),max(0,j-gap):min(j+loop,j_max)]
a = sq.im.ImageContainer(img=a_t, layer='image', lazy=True, scale=1.0, )
sq.im.segment(
img=a,
layer="image",
channel=None,
method=stardist_2D_versatile_he,
layer_added="segmented_stardist",
prob_thresh=0.01,
nms_thresh=None,
)
fig, axes = plt.subplots(1, 2, figsize=(20, 10))
a.show("image", ax=axes[0])
_ = axes[0].set_title("H&H")
a.show("segmented_stardist", cmap="jet", interpolation="none", ax=axes[1])
_ = axes[1].set_title("segmentation")
segmentation_results[(i,j)] = np.array(a['segmented_stardist'][:,:,0,0])
Found model '2D_versatile_he' for 'StarDist2D'.
Loading network weights from 'weights_best.h5'.
Loading thresholds from 'thresholds.json'.
Using default values: prob_thresh=0.692478, nms_thresh=0.3.
WARNING:tensorflow:6 out of the last 6 calls to <function Model.make_predict_function.<locals>.predict_function at 0x7fb049ab14c0> triggered tf.function retracing. Tracing is expensive and the excessive number of tracings could be due to (1) creating @tf.function repeatedly in a loop, (2) passing tensors with different shapes, (3) passing Python objects instead of tensors. For (1), please define your @tf.function outside of the loop. For (2), @tf.function has reduce_retracing=True option that can avoid unnecessary retracing. For (3), please refer to https://www.tensorflow.org/guide/function#controlling_retracing and https://www.tensorflow.org/api_docs/python/tf/function for more details.
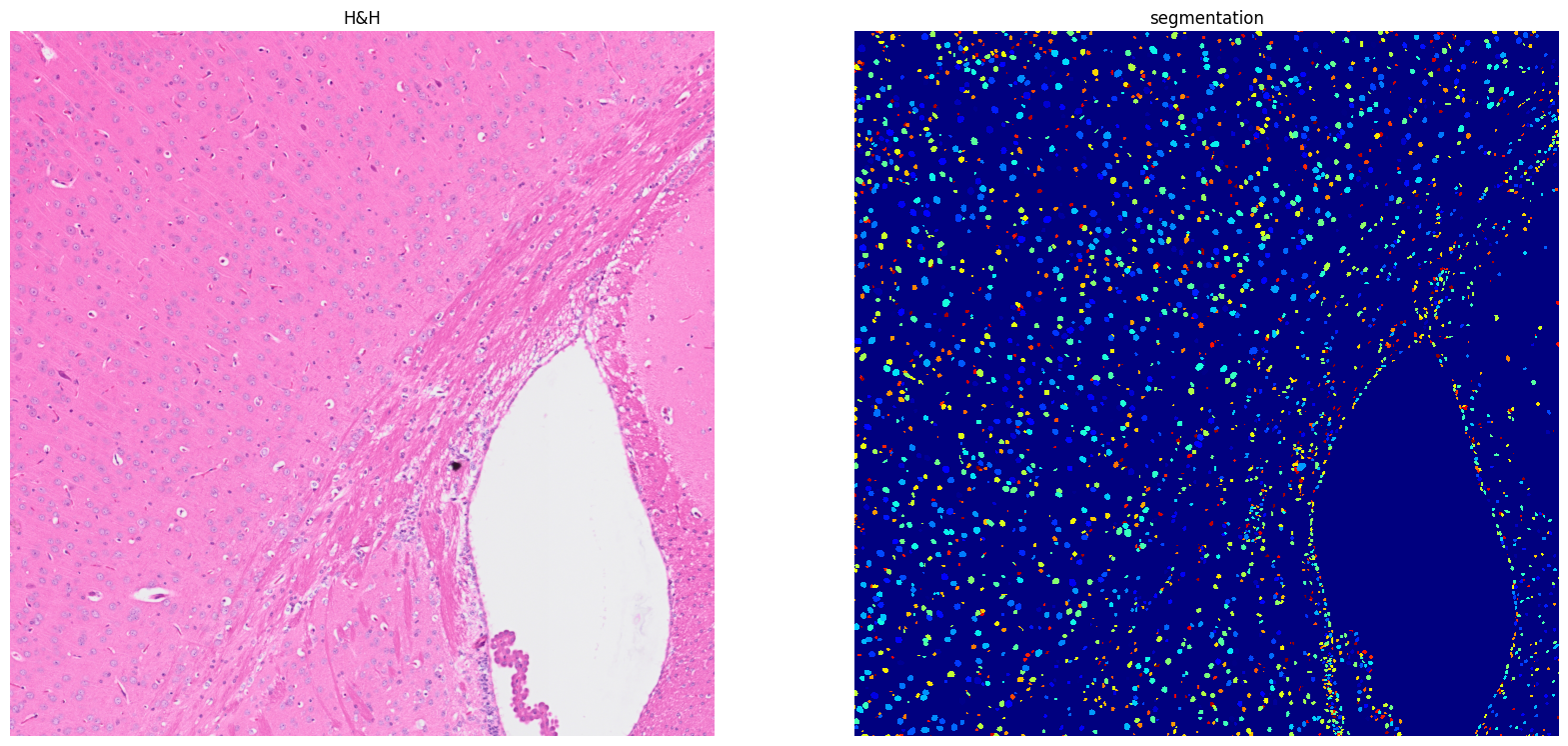
[14]:
i= loop*1
j = loop*2
a_t = image[max(0,i-gap):min(i+loop,i_max),max(0,j-gap):min(j+loop,j_max)]
a = sq.im.ImageContainer(img=a_t, layer='image', lazy=True, scale=1.0, )
sq.im.segment(
img=a,
layer="image",
channel=None,
method=stardist_2D_versatile_he,
layer_added="segmented_stardist",
prob_thresh=0.0065,
nms_thresh=None,
)
fig, axes = plt.subplots(1, 2, figsize=(20, 10))
a.show("image", ax=axes[0])
_ = axes[0].set_title("H&H")
a.show("segmented_stardist", cmap="jet", interpolation="none", ax=axes[1])
_ = axes[1].set_title("segmentation")
segmentation_results[(i,j)] = np.array(a['segmented_stardist'][:,:,0,0])
Found model '2D_versatile_he' for 'StarDist2D'.
Loading network weights from 'weights_best.h5'.
Loading thresholds from 'thresholds.json'.
Using default values: prob_thresh=0.692478, nms_thresh=0.3.
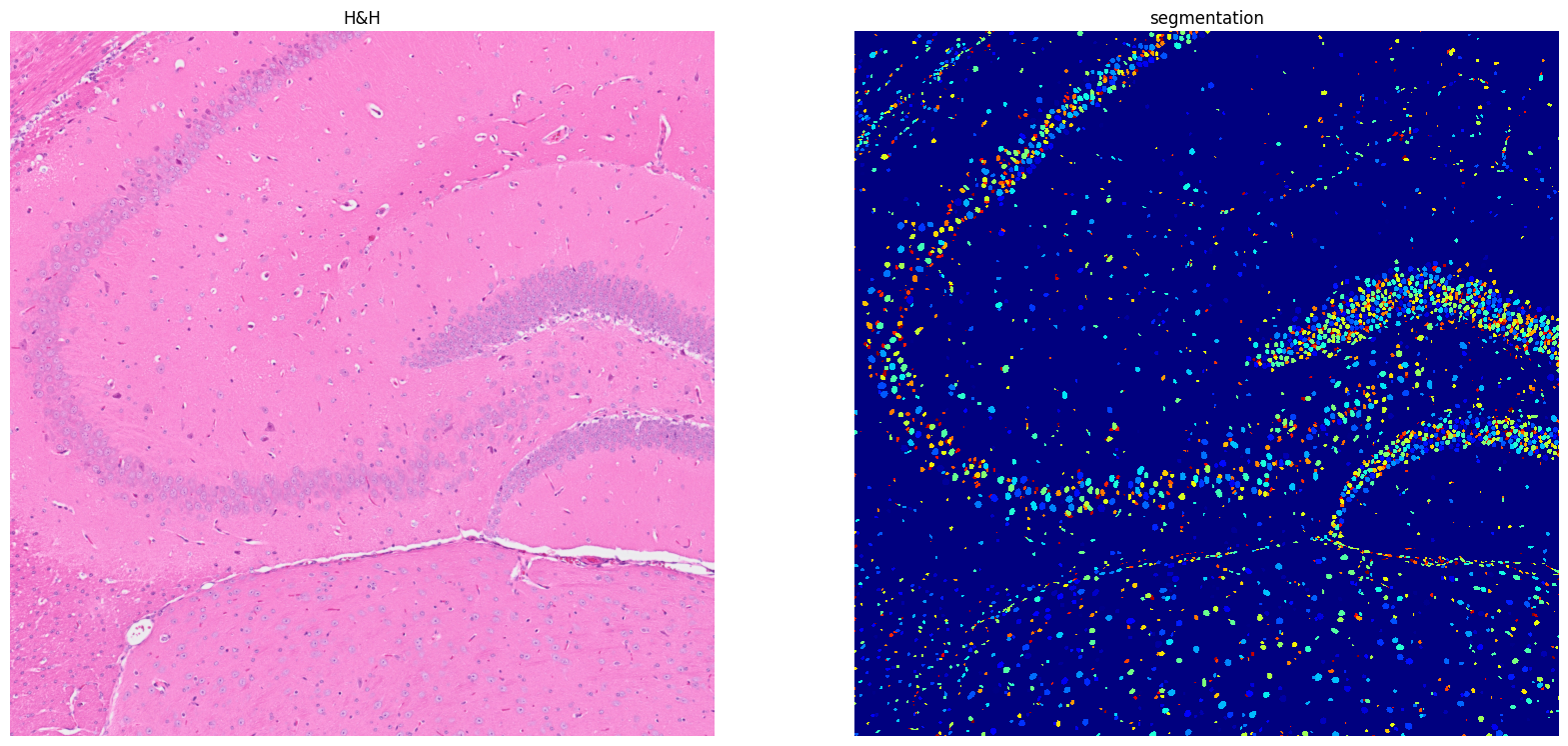
[15]:
i= loop*1
j = loop*3
a_t = image[max(0,i-gap):min(i+loop,i_max),max(0,j-gap):min(j+loop,j_max)]
a_t = clean(a_t) # No need to clean for your own data
a = sq.im.ImageContainer(img=a_t, layer='image', lazy=True, scale=1.0, )
sq.im.segment(
img=a,
layer="image",
channel=None,
method=stardist_2D_versatile_he,
layer_added="segmented_stardist",
prob_thresh=0.09,
nms_thresh=None,
)
fig, axes = plt.subplots(1, 2, figsize=(20, 10))
a.show("image", ax=axes[0])
_ = axes[0].set_title("H&H")
a.show("segmented_stardist", cmap="jet", interpolation="none", ax=axes[1])
_ = axes[1].set_title("segmentation")
segmentation_results[(i,j)] = np.array(a['segmented_stardist'][:,:,0,0])
Found model '2D_versatile_he' for 'StarDist2D'.
Loading network weights from 'weights_best.h5'.
Loading thresholds from 'thresholds.json'.
Using default values: prob_thresh=0.692478, nms_thresh=0.3.
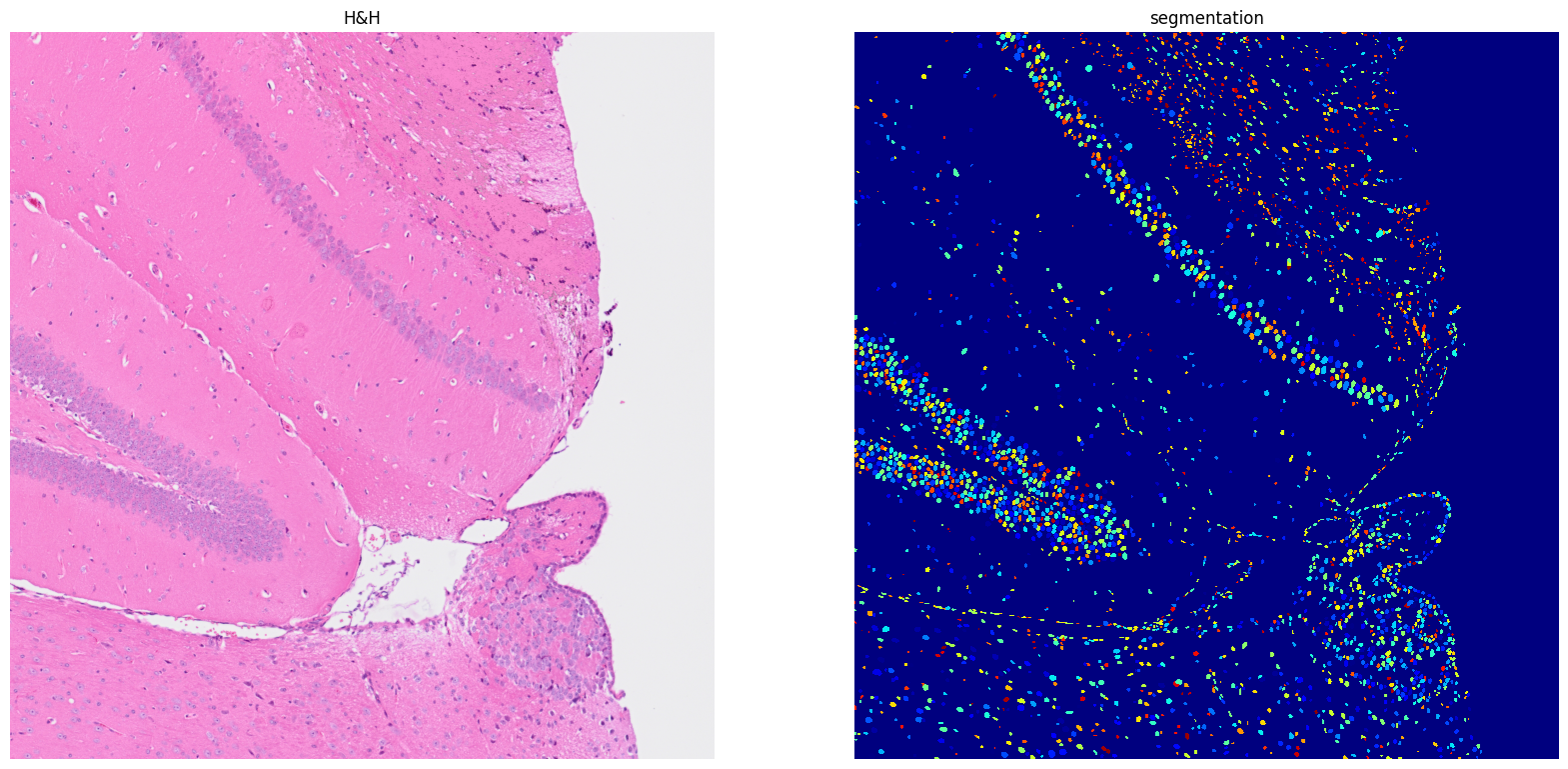
[16]:
i= loop*2
j = loop*0
a_t = image[max(0,i-gap):min(i+loop,i_max),max(0,j-gap):min(j+loop,j_max)]
a = sq.im.ImageContainer(img=a_t, layer='image', lazy=True, scale=1.0, )
sq.im.segment(
img=a,
layer="image",
channel=None,
method=stardist_2D_versatile_he,
layer_added="segmented_stardist",
prob_thresh=0.01,
nms_thresh=None,
)
fig, axes = plt.subplots(1, 2, figsize=(20, 10))
a.show("image", ax=axes[0])
_ = axes[0].set_title("H&H")
a.show("segmented_stardist", cmap="jet", interpolation="none", ax=axes[1])
_ = axes[1].set_title("segmentation")
segmentation_results[(i,j)] = np.array(a['segmented_stardist'][:,:,0,0])
Found model '2D_versatile_he' for 'StarDist2D'.
Loading network weights from 'weights_best.h5'.
Loading thresholds from 'thresholds.json'.
Using default values: prob_thresh=0.692478, nms_thresh=0.3.
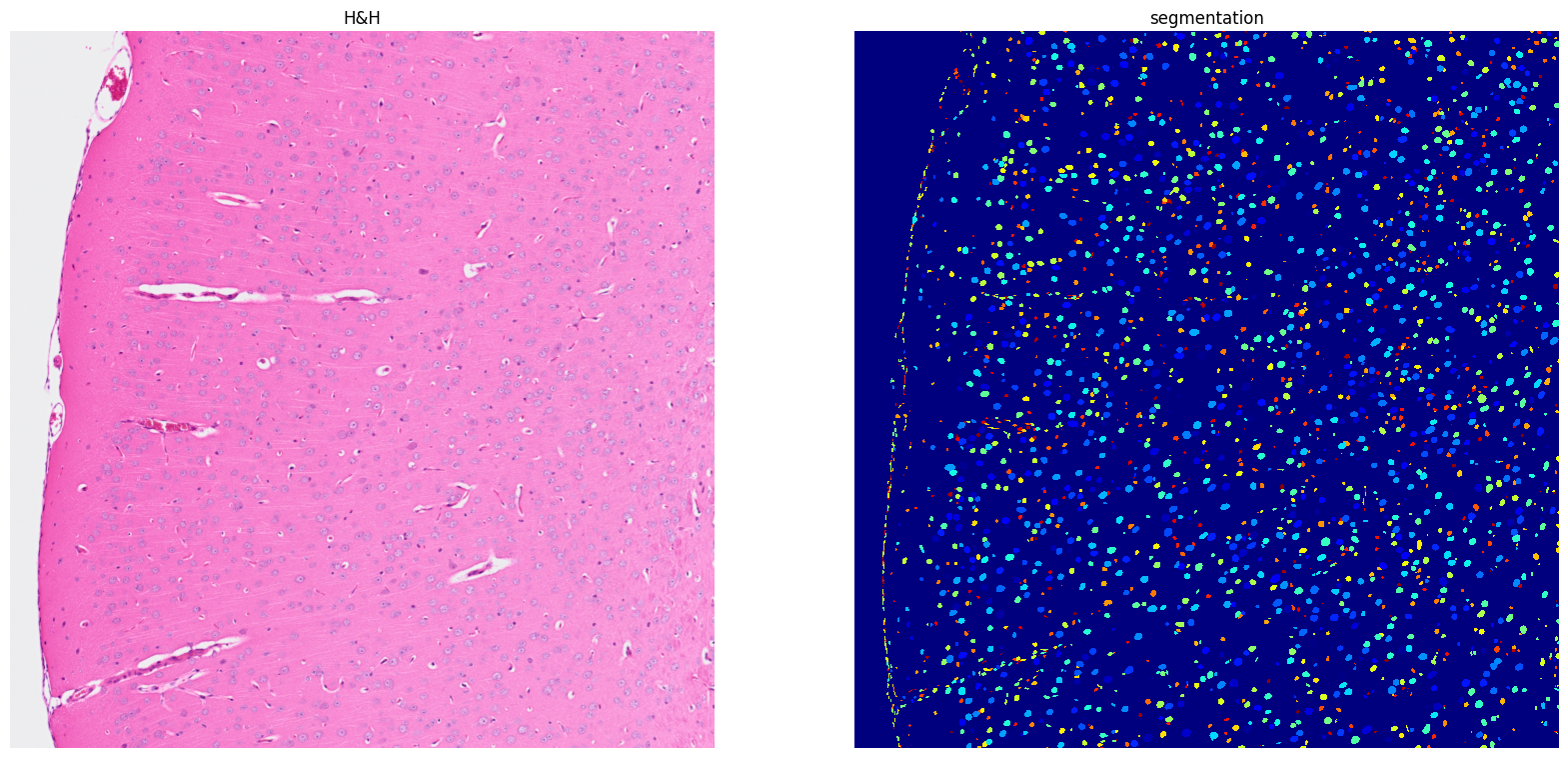
[17]:
i= loop*2
j = loop*1
a_t = image[max(0,i-gap):min(i+loop,i_max),max(0,j-gap):min(j+loop,j_max)]
a = sq.im.ImageContainer(img=a_t, layer='image', lazy=True, scale=1.0, )
sq.im.segment(
img=a,
layer="image",
channel=None,
method=stardist_2D_versatile_he,
layer_added="segmented_stardist",
prob_thresh=0.03,
nms_thresh=None,
)
fig, axes = plt.subplots(1, 2, figsize=(20, 10))
a.show("image", ax=axes[0])
_ = axes[0].set_title("H&H")
a.show("segmented_stardist", cmap="jet", interpolation="none", ax=axes[1])
_ = axes[1].set_title("segmentation")
segmentation_results[(i,j)] = np.array(a['segmented_stardist'][:,:,0,0])
Found model '2D_versatile_he' for 'StarDist2D'.
Loading network weights from 'weights_best.h5'.
Loading thresholds from 'thresholds.json'.
Using default values: prob_thresh=0.692478, nms_thresh=0.3.
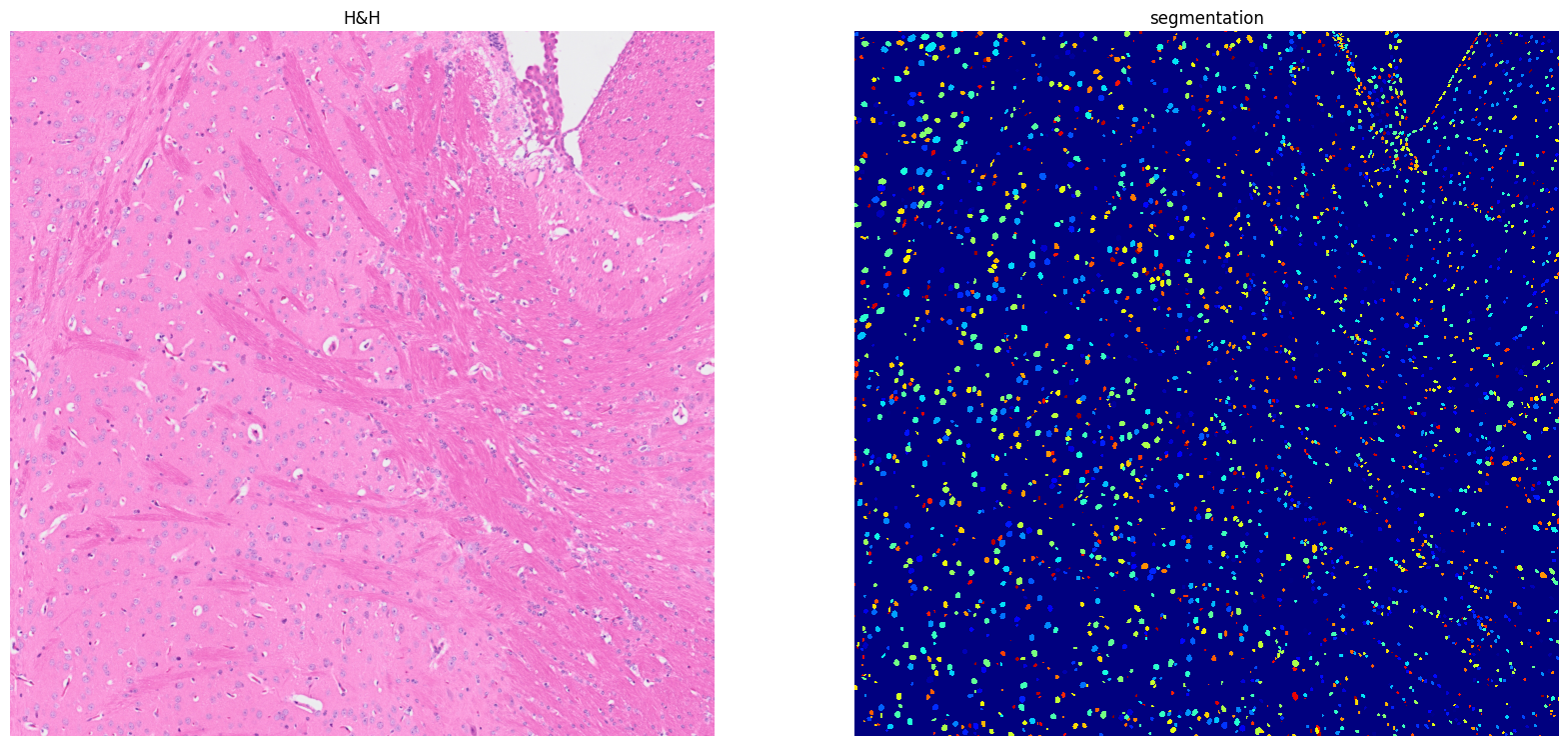
[18]:
i= loop*2
j = loop*2
a_t = image[max(0,i-gap):min(i+loop,i_max),max(0,j-gap):min(j+loop,j_max)]
a = sq.im.ImageContainer(img=a_t, layer='image', lazy=True, scale=1.0, )
sq.im.segment(
img=a,
layer="image",
channel=None,
method=stardist_2D_versatile_he,
layer_added="segmented_stardist",
prob_thresh=0.01,
nms_thresh=None,
)
fig, axes = plt.subplots(1, 2, figsize=(20, 10))
a.show("image", ax=axes[0])
_ = axes[0].set_title("H&H")
a.show("segmented_stardist", cmap="jet", interpolation="none", ax=axes[1])
_ = axes[1].set_title("segmentation")
segmentation_results[(i,j)] = np.array(a['segmented_stardist'][:,:,0,0])
Found model '2D_versatile_he' for 'StarDist2D'.
Loading network weights from 'weights_best.h5'.
Loading thresholds from 'thresholds.json'.
Using default values: prob_thresh=0.692478, nms_thresh=0.3.
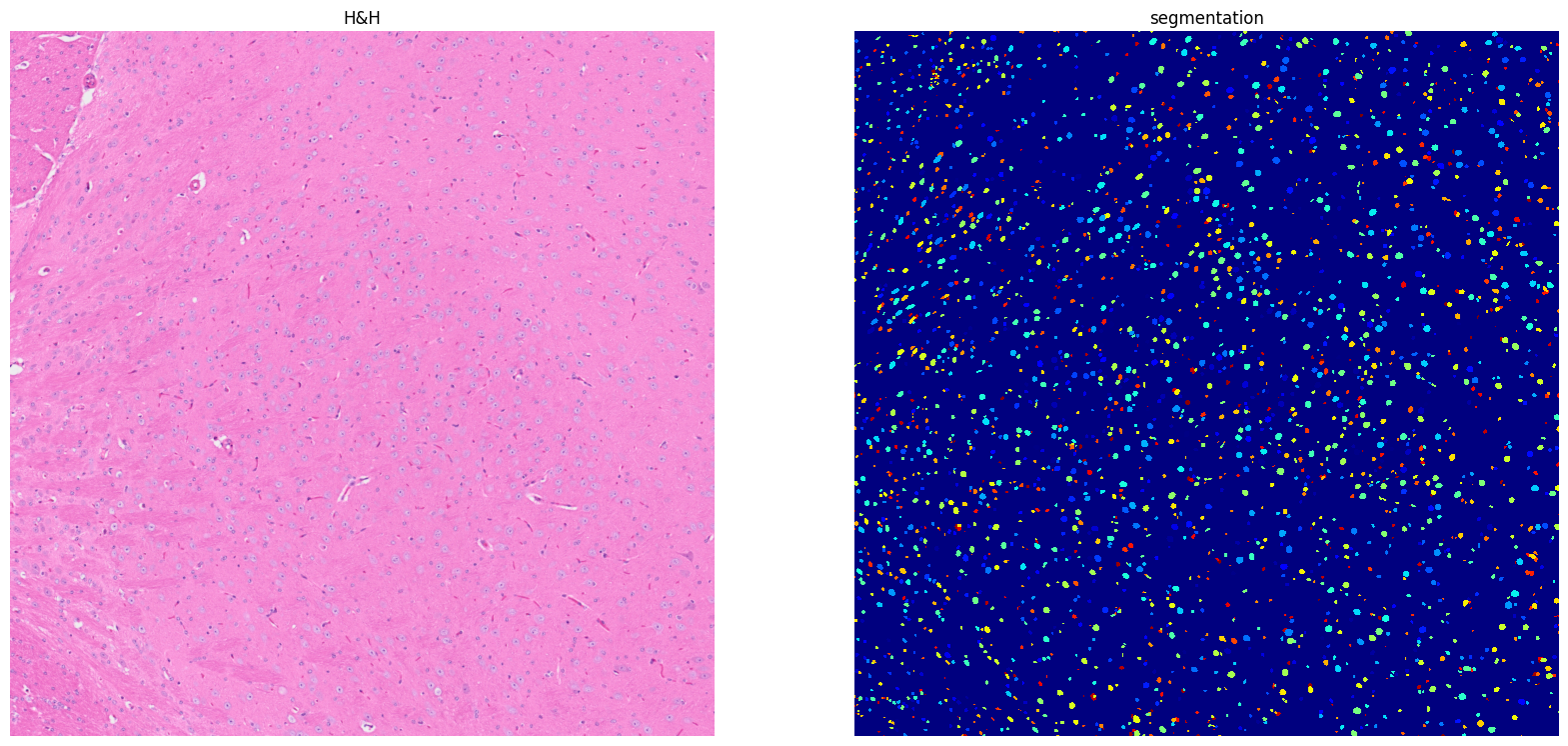
[19]:
i= loop*2
j = loop*3
a_t = image[max(0,i-gap):min(i+loop,i_max),max(0,j-gap):min(j+loop,j_max)]
a = sq.im.ImageContainer(img=a_t, layer='image', lazy=True, scale=1.0, )
sq.im.segment(
img=a,
layer="image",
channel=None,
method=stardist_2D_versatile_he,
layer_added="segmented_stardist",
prob_thresh=0.01,
nms_thresh=None,
)
fig, axes = plt.subplots(1, 2, figsize=(20, 10))
a.show("image", ax=axes[0])
_ = axes[0].set_title("H&H")
a.show("segmented_stardist", cmap="jet", interpolation="none", ax=axes[1])
_ = axes[1].set_title("segmentation")
segmentation_results[(i,j)] = np.array(a['segmented_stardist'][:,:,0,0])
Found model '2D_versatile_he' for 'StarDist2D'.
Loading network weights from 'weights_best.h5'.
Loading thresholds from 'thresholds.json'.
Using default values: prob_thresh=0.692478, nms_thresh=0.3.
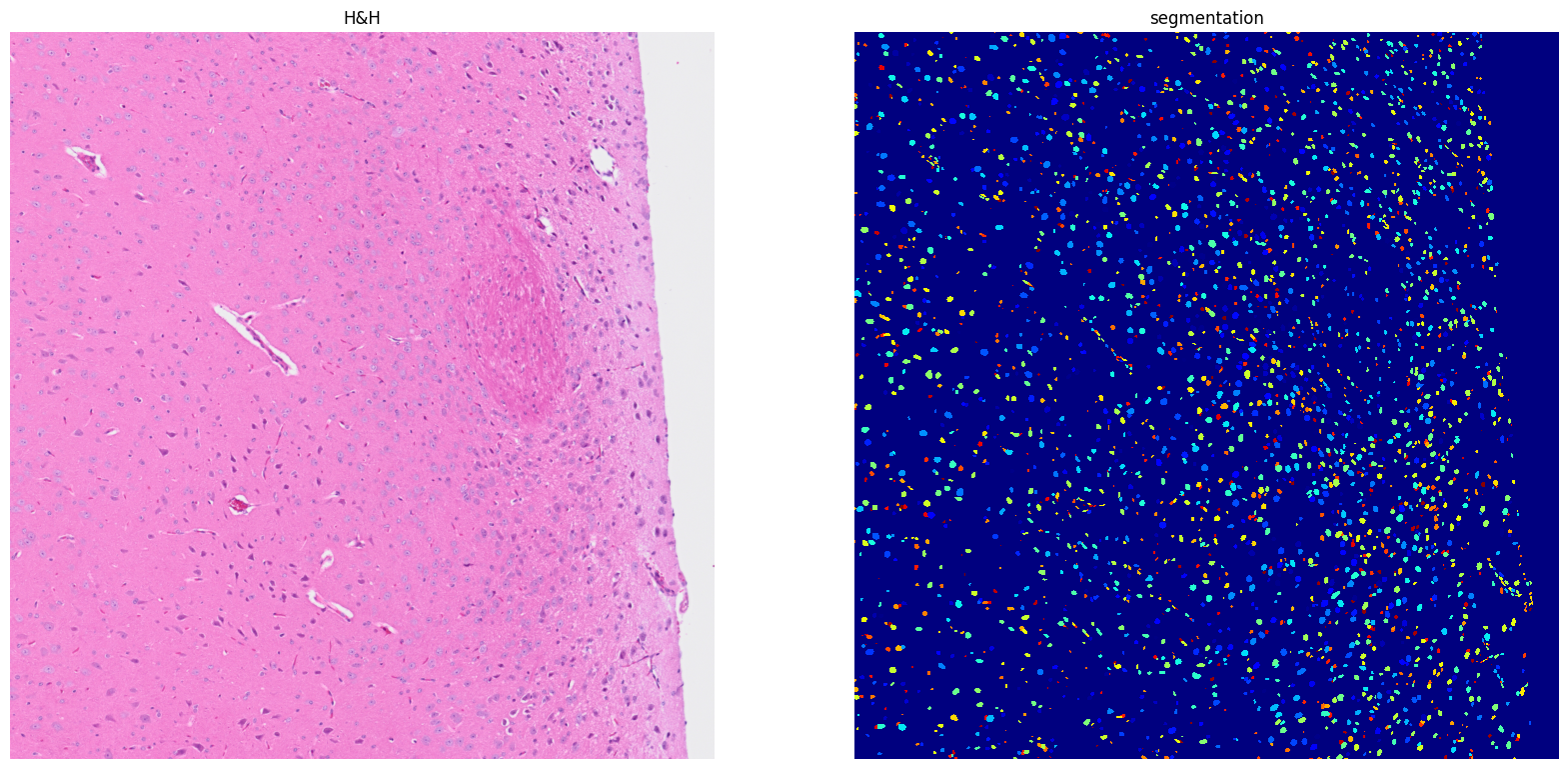
[20]:
i= loop*3
j = loop*0
a_t = image[max(0,i-gap):min(i+loop,i_max),max(0,j-gap):min(j+loop,j_max)]
a = sq.im.ImageContainer(img=a_t, layer='image', lazy=True, scale=1.0, )
sq.im.segment(
img=a,
layer="image",
channel=None,
method=stardist_2D_versatile_he,
layer_added="segmented_stardist",
prob_thresh=0.01,
nms_thresh=None,
)
fig, axes = plt.subplots(1, 2, figsize=(20, 10))
a.show("image", ax=axes[0])
_ = axes[0].set_title("H&H")
a.show("segmented_stardist", cmap="jet", interpolation="none", ax=axes[1])
_ = axes[1].set_title("segmentation")
segmentation_results[(i,j)] = np.array(a['segmented_stardist'][:,:,0,0])
Found model '2D_versatile_he' for 'StarDist2D'.
Loading network weights from 'weights_best.h5'.
Loading thresholds from 'thresholds.json'.
Using default values: prob_thresh=0.692478, nms_thresh=0.3.
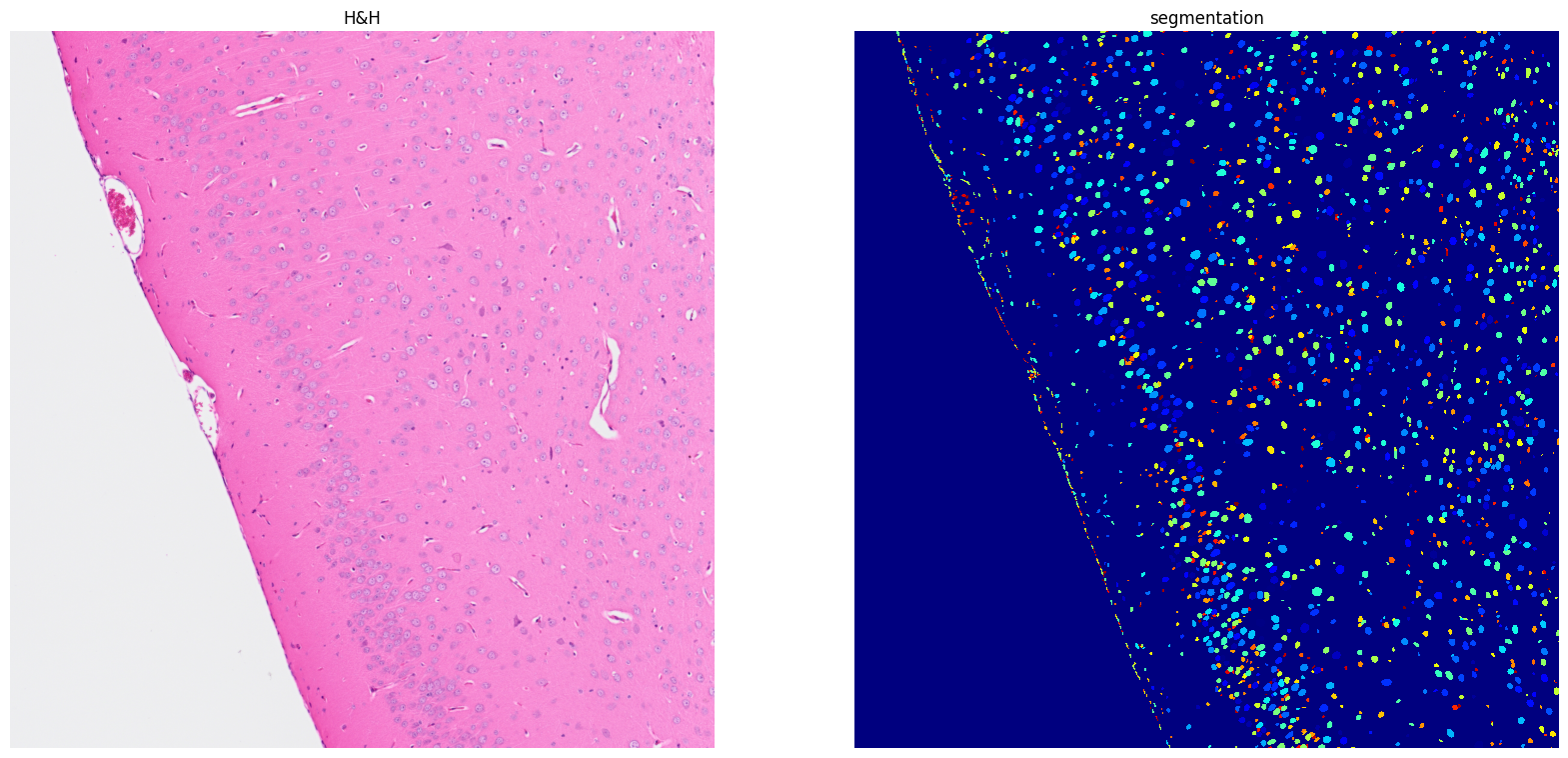
[21]:
i= loop*3
j = loop*1
a_t = image[max(0,i-gap):min(i+loop,i_max),max(0,j-gap):min(j+loop,j_max)]
a = sq.im.ImageContainer(img=a_t, layer='image', lazy=True, scale=1.0, )
sq.im.segment(
img=a,
layer="image",
channel=None,
method=stardist_2D_versatile_he,
layer_added="segmented_stardist",
prob_thresh=0.01,
nms_thresh=None,
)
fig, axes = plt.subplots(1, 2, figsize=(20, 10))
a.show("image", ax=axes[0])
_ = axes[0].set_title("H&H")
a.show("segmented_stardist", cmap="jet", interpolation="none", ax=axes[1])
_ = axes[1].set_title("segmentation")
segmentation_results[(i,j)] = np.array(a['segmented_stardist'][:,:,0,0])
Found model '2D_versatile_he' for 'StarDist2D'.
Loading network weights from 'weights_best.h5'.
Loading thresholds from 'thresholds.json'.
Using default values: prob_thresh=0.692478, nms_thresh=0.3.
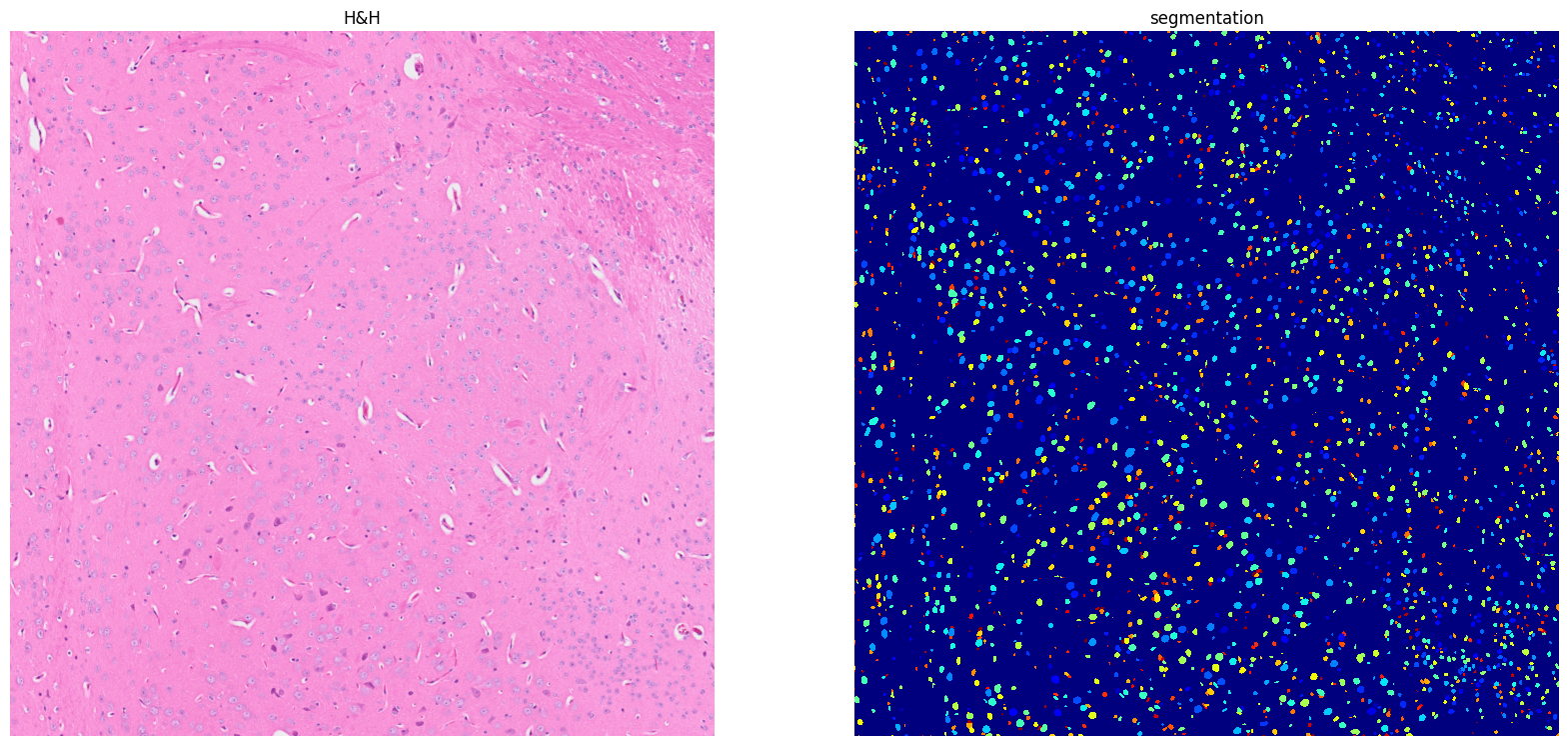
[22]:
i= loop*3
j = loop*2
a_t = image[max(0,i-gap):min(i+loop,i_max),max(0,j-gap):min(j+loop,j_max)]
a = sq.im.ImageContainer(img=a_t, layer='image', lazy=True, scale=1.0, )
sq.im.segment(
img=a,
layer="image",
channel=None,
method=stardist_2D_versatile_he,
layer_added="segmented_stardist",
prob_thresh=0.01,
nms_thresh=None,
)
fig, axes = plt.subplots(1, 2, figsize=(20, 10))
a.show("image", ax=axes[0])
_ = axes[0].set_title("H&H")
a.show("segmented_stardist", cmap="jet", interpolation="none", ax=axes[1])
_ = axes[1].set_title("segmentation")
segmentation_results[(i,j)] = np.array(a['segmented_stardist'][:,:,0,0])
Found model '2D_versatile_he' for 'StarDist2D'.
Loading network weights from 'weights_best.h5'.
Loading thresholds from 'thresholds.json'.
Using default values: prob_thresh=0.692478, nms_thresh=0.3.
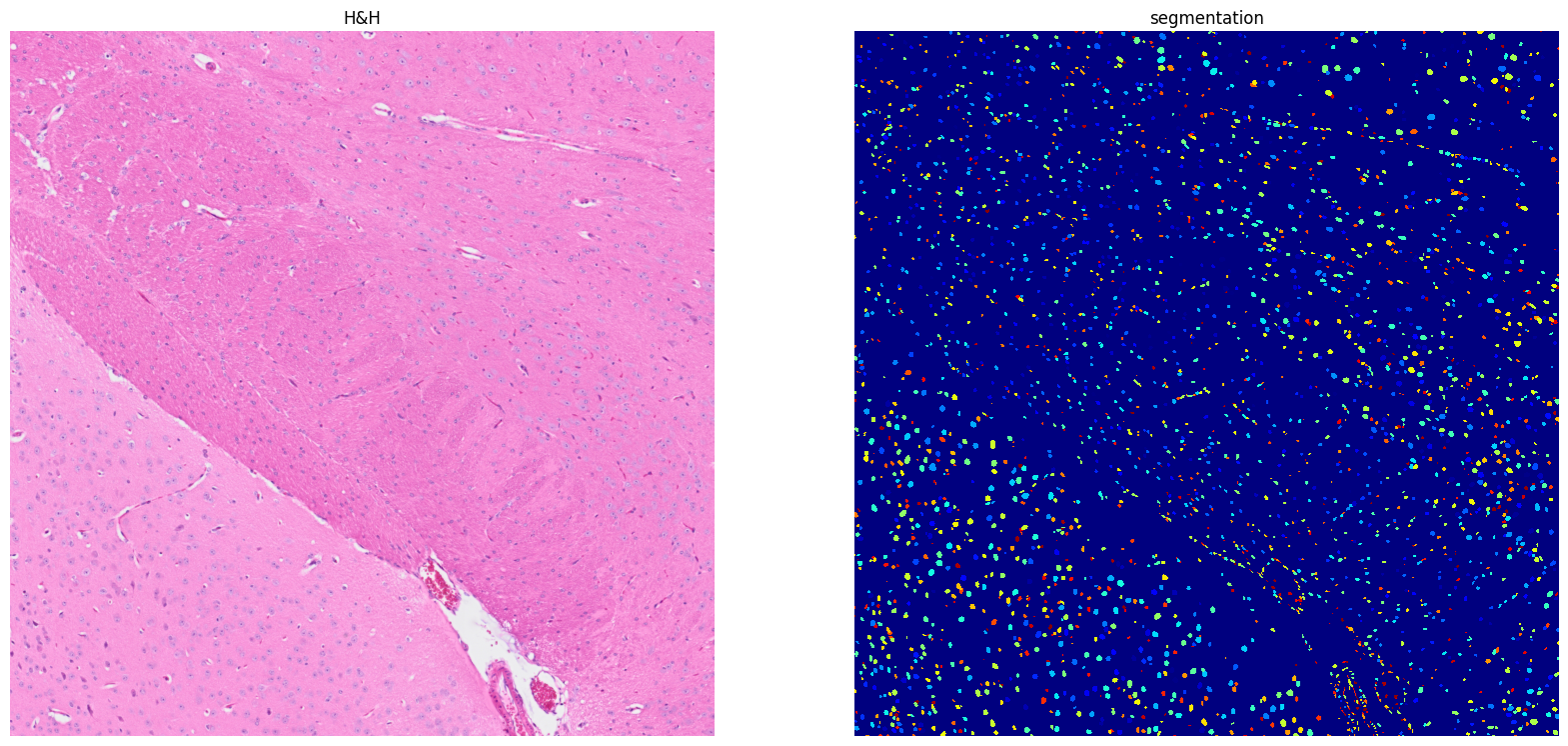
[23]:
i= loop*3
j = loop*3
a_t = image[max(0,i-gap):min(i+loop,i_max),max(0,j-gap):min(j+loop,j_max)]
a = sq.im.ImageContainer(img=a_t, layer='image', lazy=True, scale=1.0, )
sq.im.segment(
img=a,
layer="image",
channel=None,
method=stardist_2D_versatile_he,
layer_added="segmented_stardist",
prob_thresh=0.01,
nms_thresh=None,
)
fig, axes = plt.subplots(1, 2, figsize=(20, 10))
a.show("image", ax=axes[0])
_ = axes[0].set_title("H&H")
a.show("segmented_stardist", cmap="jet", interpolation="none", ax=axes[1])
_ = axes[1].set_title("segmentation")
segmentation_results[(i,j)] = np.array(a['segmented_stardist'][:,:,0,0])
Found model '2D_versatile_he' for 'StarDist2D'.
Loading network weights from 'weights_best.h5'.
Loading thresholds from 'thresholds.json'.
Using default values: prob_thresh=0.692478, nms_thresh=0.3.
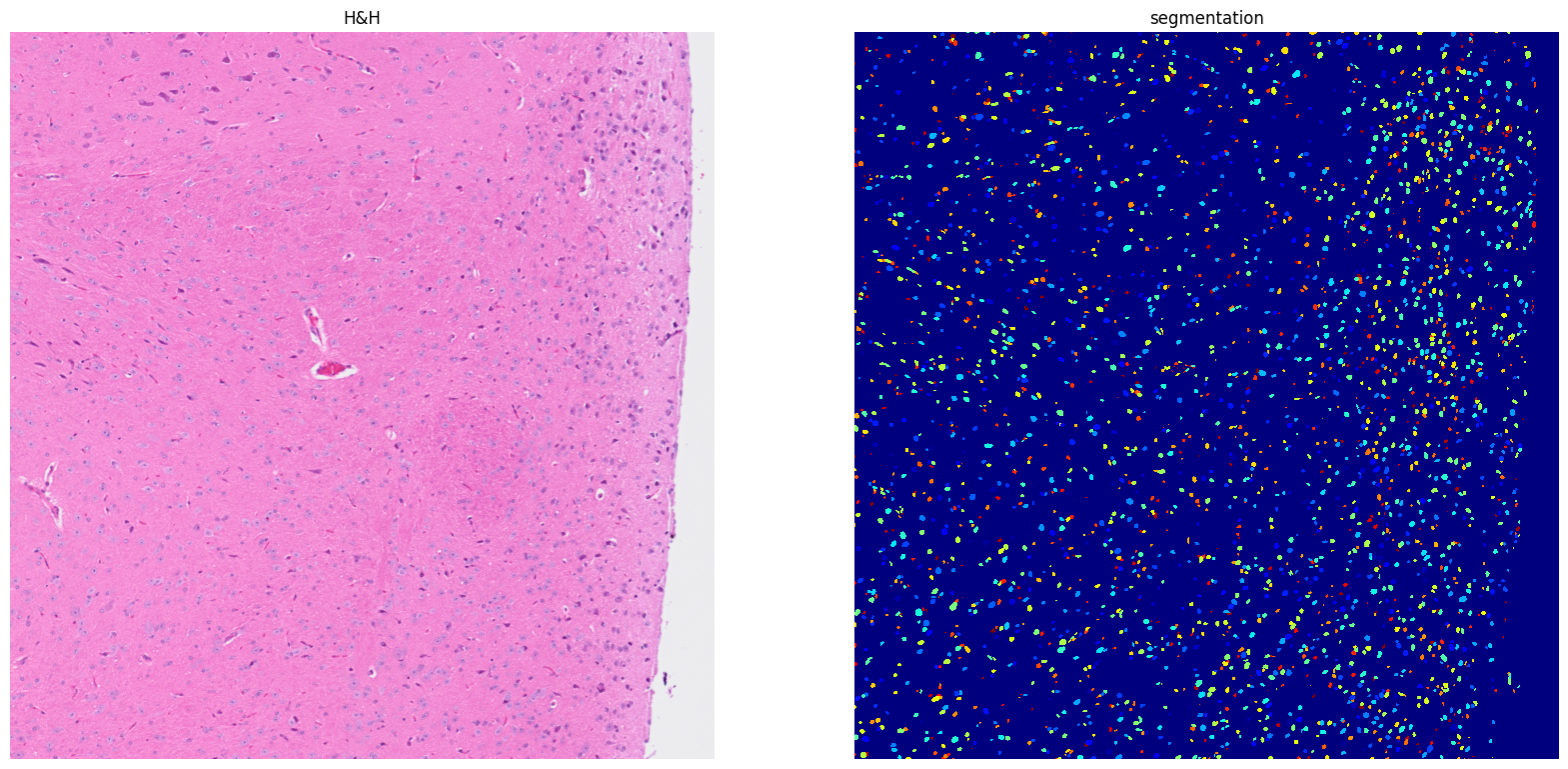
[24]:
i= loop*4
j = loop*0
a_t = image[max(0,i-gap):min(i+loop,i_max),max(0,j-gap):min(j+loop,j_max)]
a = sq.im.ImageContainer(img=a_t, layer='image', lazy=True, scale=1.0, )
sq.im.segment(
img=a,
layer="image",
channel=None,
method=stardist_2D_versatile_he,
layer_added="segmented_stardist",
prob_thresh=0.01,
nms_thresh=None,
)
fig, axes = plt.subplots(1, 2, figsize=(20, 10))
a.show("image", ax=axes[0])
_ = axes[0].set_title("H&H")
a.show("segmented_stardist", cmap="jet", interpolation="none", ax=axes[1])
_ = axes[1].set_title("segmentation")
segmentation_results[(i,j)] = np.array(a['segmented_stardist'][:,:,0,0])
Found model '2D_versatile_he' for 'StarDist2D'.
Loading network weights from 'weights_best.h5'.
Loading thresholds from 'thresholds.json'.
Using default values: prob_thresh=0.692478, nms_thresh=0.3.

[25]:
i= loop*4
j = loop*1
a_t = image[max(0,i-gap):min(i+loop,i_max),max(0,j-gap):min(j+loop,j_max)]
a = sq.im.ImageContainer(img=a_t, layer='image', lazy=True, scale=1.0, )
sq.im.segment(
img=a,
layer="image",
channel=None,
method=stardist_2D_versatile_he,
layer_added="segmented_stardist",
prob_thresh=0.01,
nms_thresh=None,
)
fig, axes = plt.subplots(1, 2, figsize=(20, 10))
a.show("image", ax=axes[0])
_ = axes[0].set_title("H&H")
a.show("segmented_stardist", cmap="jet", interpolation="none", ax=axes[1])
_ = axes[1].set_title("segmentation")
segmentation_results[(i,j)] = np.array(a['segmented_stardist'][:,:,0,0])
Found model '2D_versatile_he' for 'StarDist2D'.
Loading network weights from 'weights_best.h5'.
Loading thresholds from 'thresholds.json'.
Using default values: prob_thresh=0.692478, nms_thresh=0.3.
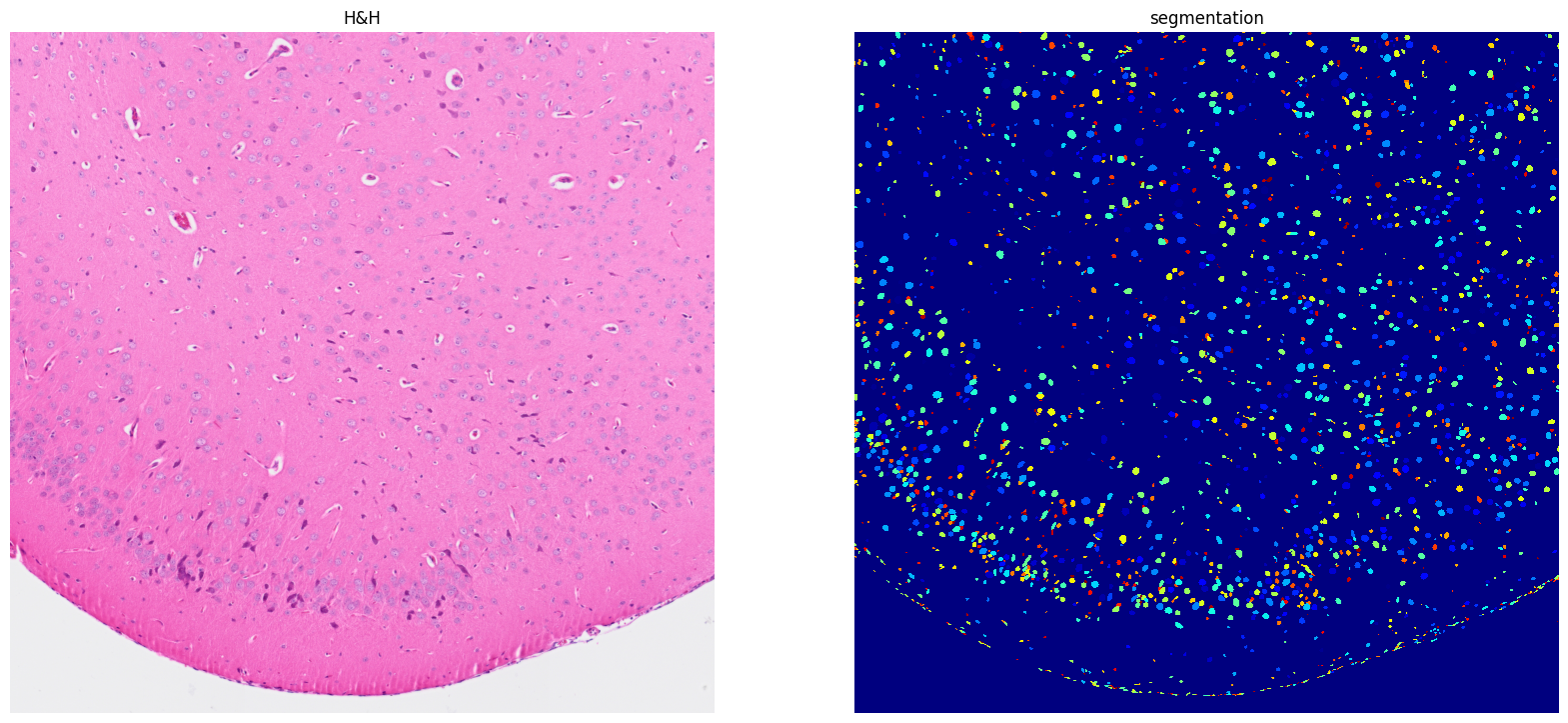
[26]:
i= loop*4
j = loop*2
a_t = image[max(0,i-gap):min(i+loop,i_max),max(0,j-gap):min(j+loop,j_max)]
a = sq.im.ImageContainer(img=a_t, layer='image', lazy=True, scale=1.0, )
sq.im.segment(
img=a,
layer="image",
channel=None,
method=stardist_2D_versatile_he,
layer_added="segmented_stardist",
prob_thresh=0.01,
nms_thresh=None,
)
fig, axes = plt.subplots(1, 2, figsize=(20, 10))
a.show("image", ax=axes[0])
_ = axes[0].set_title("H&H")
a.show("segmented_stardist", cmap="jet", interpolation="none", ax=axes[1])
_ = axes[1].set_title("segmentation")
segmentation_results[(i,j)] = np.array(a['segmented_stardist'][:,:,0,0])
Found model '2D_versatile_he' for 'StarDist2D'.
Loading network weights from 'weights_best.h5'.
Loading thresholds from 'thresholds.json'.
Using default values: prob_thresh=0.692478, nms_thresh=0.3.
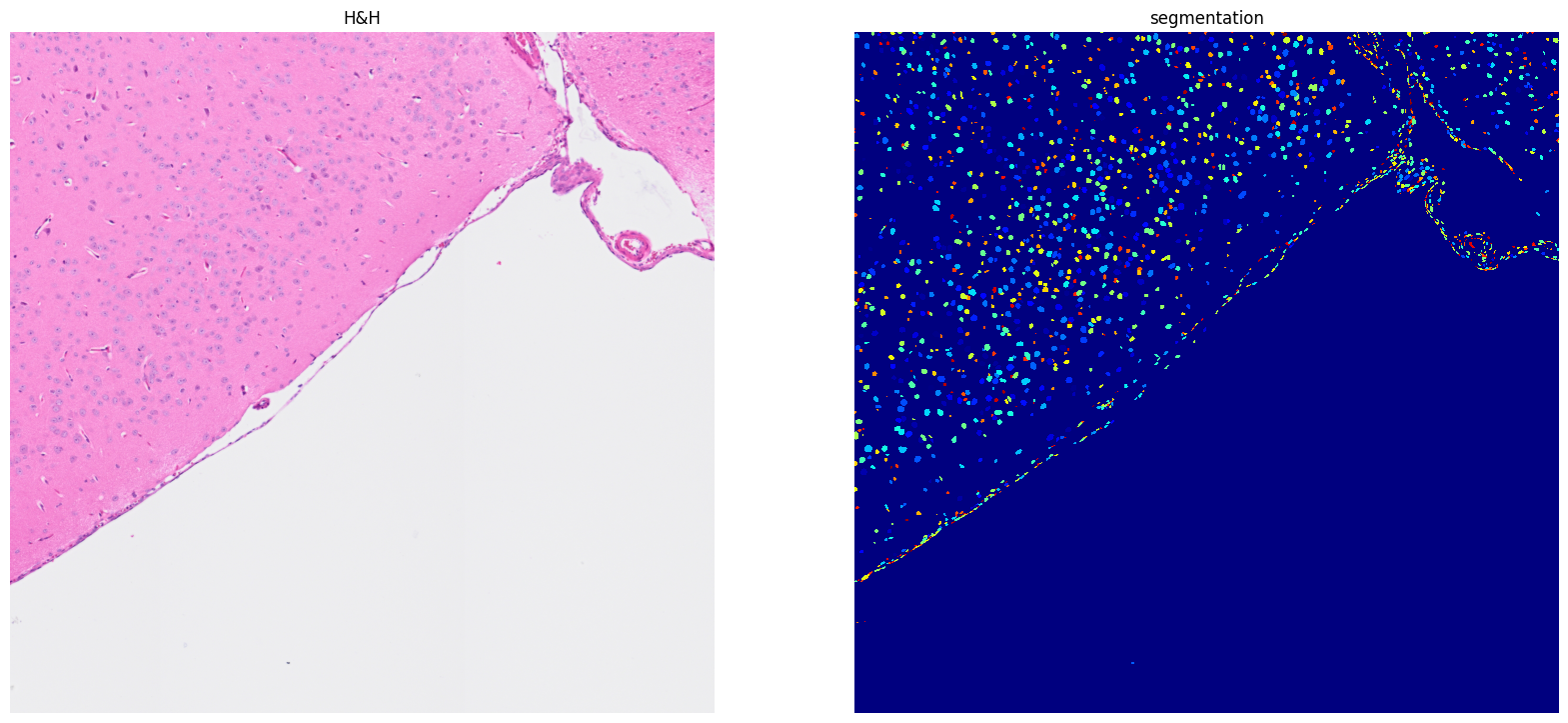
[27]:
i= loop*4
j = loop*3
a_t = image[max(0,i-gap):min(i+loop,i_max),max(0,j-gap):min(j+loop,j_max)]
a = sq.im.ImageContainer(img=a_t, layer='image', lazy=True, scale=1.0, )
sq.im.segment(
img=a,
layer="image",
channel=None,
method=stardist_2D_versatile_he,
layer_added="segmented_stardist",
prob_thresh=0.01,
nms_thresh=None,
)
fig, axes = plt.subplots(1, 2, figsize=(20, 10))
a.show("image", ax=axes[0])
_ = axes[0].set_title("H&H")
a.show("segmented_stardist", cmap="jet", interpolation="none", ax=axes[1])
_ = axes[1].set_title("segmentation")
segmentation_results[(i,j)] = np.array(a['segmented_stardist'][:,:,0,0])
Found model '2D_versatile_he' for 'StarDist2D'.
Loading network weights from 'weights_best.h5'.
Loading thresholds from 'thresholds.json'.
Using default values: prob_thresh=0.692478, nms_thresh=0.3.
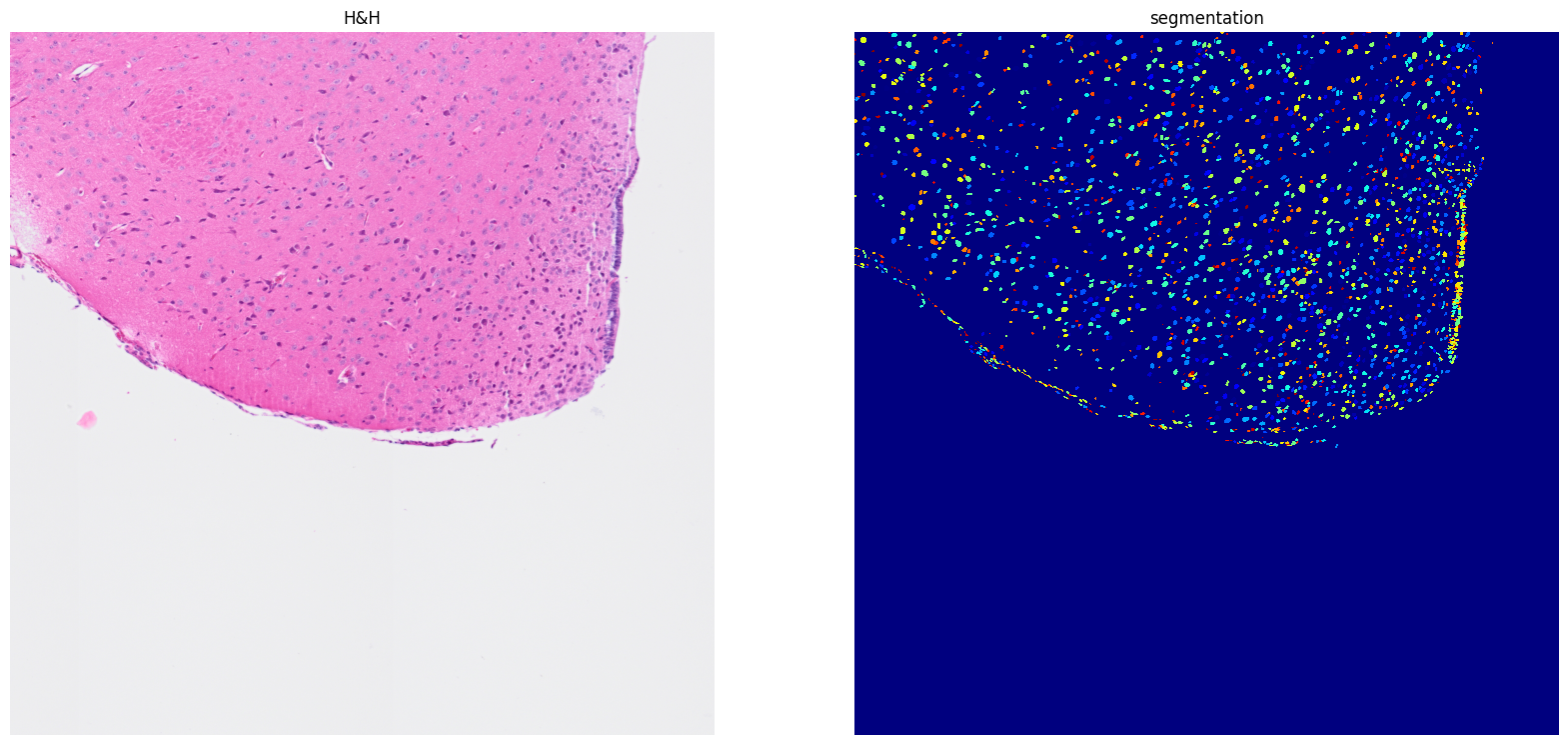
Here, we have a function to combine all the smaller segments into the final complete image.
[28]:
def segmentation_final(segmentation_results, i_max, j_max, loop, gap):
# Combine segmentation results from different blocks into a final segmentation map
segmentation_results1 = copy.deepcopy(segmentation_results)
num = 0
# Adjust labels to ensure uniqueness across blocks
for i in range(0,i_max,loop):
for j in range(0,j_max,loop):
segmentation_results1[(i,j)][segmentation_results1[(i,j)] != 0] = segmentation_results1[(i,j)][segmentation_results1[(i,j)] != 0] + num
num = max(num,segmentation_results1[(i,j)].max())
segmentation_final = np.zeros((i_max, j_max))
# Merge the blocks into the final segmentation map
for i in range(0,i_max,loop):
for j in range(0,j_max,loop):
if i == 0 and j == 0:
segmentation_final[i:min(i+loop,i_max),j:min(j+loop,j_max)] = segmentation_results1[(i,j)]
elif i == 0 and j != 0:
segmentation_final[i:min(i+loop,i_max),j:min(j+loop,j_max)] = segmentation_results1[(i,j)][:,gap:]
elif i != 0 and j == 0:
segmentation_final[i:min(i+loop,i_max),j:min(j+loop,j_max)] = segmentation_results1[(i,j)][gap:,:]
else:
segmentation_final[i:min(i+loop,i_max),j:min(j+loop,j_max)] = segmentation_results1[(i,j)][gap:,gap:]
t = 0
# Resolve overlaps between blocks
for i in range(0,i_max,loop):
for j in range(0,j_max,loop):
if j!= 0:
if i == 0:
a = copy.deepcopy(segmentation_results1[(i,(j-loop))][:,-gap:])
b = copy.deepcopy(segmentation_results1[(i,j)][:,:gap])
uni_a = np.unique(a[:,-1:])
uni_a = uni_a[uni_a > 0]
for ele in uni_a:
uni_b = np.unique(b[a == ele])
uni_b = uni_b[uni_b>0]
if len(uni_b) > 0:
a[a == ele] =0
for eleb in uni_b:
a[b == eleb] = eleb
segmentation_final[i:min(i+loop,i_max),min(j,j_max)-gap:min(j,j_max)] = copy.deepcopy(a)
else:
a = copy.deepcopy(segmentation_results1[(i,(j-loop))][gap:,-gap:])
b = copy.deepcopy(segmentation_results1[(i,j)][gap:,:gap])
uni_a = np.unique(a[:,-1:])
uni_a = uni_a[uni_a > 0]
for ele in uni_a:
uni_b = np.unique(b[a == ele])
uni_b = uni_b[uni_b>0]
if len(uni_b) > 0:
t= t+1
a[a == ele] =0
for eleb in uni_b:
a[b == eleb] = eleb
segmentation_final[i:min(i+loop,i_max),min(j,j_max)-gap:min(j,j_max)] = copy.deepcopy(a)
if i!= 0:
if j == 0:
a = copy.deepcopy(segmentation_results1[((i-loop),j)][-gap:,:])
b = copy.deepcopy(segmentation_results1[(i,j)][:gap,:])
uni_a = np.unique(a[-1:,:])
uni_a = uni_a[uni_a > 0]
for ele in uni_a:
uni_b = np.unique(b[a == ele])
uni_b = uni_b[uni_b>0]
if len(uni_b) > 0:
t= t+1
a[a == ele] =0
for eleb in uni_b:
a[b == eleb] = eleb
segmentation_final[(min(i,i_max)-gap):min(i,i_max),j:min(j+loop,j_max)] = copy.deepcopy(a)
else:
a = copy.deepcopy(segmentation_results1[((i-loop),j)][-gap:,gap:])
b = copy.deepcopy(segmentation_results1[(i,j)][:gap,gap:])
uni_a = np.unique(a[-1:,:])
uni_a = uni_a[uni_a > 0]
for ele in uni_a:
uni_b = np.unique(b[a == ele])
uni_b = uni_b[uni_b>0]
if len(uni_b) > 0:
t= t+1
a[a == ele] =0
for eleb in uni_b:
a[b == eleb] = eleb
segmentation_final[(min(i,i_max)-gap):min(i,i_max),j:min(j+loop,j_max)] = copy.deepcopy(a)
return segmentation_final
segmentation_final = segmentation_final(segmentation_results, i_max, j_max, loop, gap)
Save result here.
[ ]:
np.save(data_path + 'segmentation_final.npy', segmentation_final)